The G-Q-reat ESKAPE project funded by the Marie Skłodowska-Curie actions
The World Health Organization has identified antimicrobial resistance as one of the top three health threats of the 21 st century, and it has recently published a priority list of multi-drug resistance bacteria for which new antibiotics are urgently needed. Noteworthy, the WHO has underlined the criticisms and risks associated to the current antimicrobial resistance highlighting that we are potentially exposed to a bacteria outbreak as emphasized by the COVID-19 pandemic.

Stefano Ciaco, Marie Curie postdoctoral researcher at University of Siena, spoke to us about the multidisciplinary project G-Q-reat ESKAPE, funded by the Marie Skłodowska-Curie actions.
The focus of the project is to contrast the antimicrobial resistance using a multidisciplinary approach combing in silico approaches, such as molecular dynamic simulations and docking, and biophysical tools, such as thermal shift assay, circular dichroism and fluorescent-based studies.
“For decades – says Ciaco – antimicrobial resistance was underestimated and fueled by the misuse and massive use of antibiotics for prophylaxis therapies and inappropriate self-medication in humans, food-producing animal prophylaxis and release of antibiotics in wastewaters. Moreover, very few attempts were made to discover and develop new clinically relevant antibiotics, mostly of them are close chemical derivatives of existing antimicrobial drugs, which represents a short-term solution as the antimicrobial resistance was already established. As a result, bacterial infections are representing a concrete health threat worldwide, with the potential to become one of the major causes of death in a few years”.
World Health Organization identified six highly virulent and pan-drug resistant pathogens that are commonly classified with the acronym E.S.K.A.P.E., which includes Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and, Enterobacter spp..
Other bacteria included in the World Health Organization priority list are Escherichia coli, Helicobacter pylori, Campylobacter spp., Salmonellae, Neisseria gonorrhoeae, Streptococcus pneumoniae, Haemophilus influenzae, Shigella spp..

“The G-Q-reat ESKAPE project – Ciaco explains – aims to target Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii and, Pseudomonas aeruginosa, which are representative members of Gram-negative bacteria. In addition, Staphylococcus aureus and Streptococcus pneumoniae are targeted as representative members of Gram-positive bacteria. Overall, due to the high rate of mutations of the pathogens, G-quadruplex structures represent a favorable and potential target”.
“G-quadruplexes – Ciaco continues – are non-canonical four-stranded structural motifs, identified both in DNA and RNA, formed by guanine-rich sequences playing a wide range of biological functions in human and non-human cells. The fundamental unit of G-quadruplex structures is the tetrad which is composed of four co-planar guanine nucleotides held together via Hoogsteen hydrogen bonds and coordinated by positively charged ions such as potassium or sodium, multiple tetrads stacked on each other form the G-quadruplex structure. Originally, the G-quadruplex sequences were identified in human telomers and oncogene promoter regions playing a critical role in telomer shortening (cell aging) and transcription regulation (cancer development), respectively. Indeed, G-quadruplex binders were investigated as potential anticancer activity in humans”.
“Several potential G-quadruplex sequences – says Ciaco – were found in other organisms such as vertebrates (evolutionary close to Homo sapiens), yeasts (i.e. Saccharomyces cerevisiae), viruses (i.e. human immunodeficiency virus (HIV-1), parasites (i.e. Plasmodium falciparum) and bacteria (i.e. Escherichia coli). Nevertheless, the critical role of G-quadruplexes in bacteria replication, metabolism, synthesis of secondary metabolites (such as toxins and biofilms), virulence and drug- resistant genes were not yet fully elucidated. Furthermore, G-quadruplexes are highly conserved in different bacteria and along the same strains thus, GQ binders are potentially broad- spectrum antibiotics”.
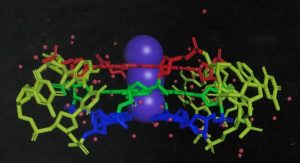
“Overall – Ciaco concludes – the G-Q-reat ESKAPE project aims to gain knowledge on bacteria G-quadruplex structure and folding by a multidisciplinary approach combining computational rational design of G-quadruplex selective binders with biophysical studies. Another goal is to optime specific modulators of bacteria G-quadruplexes endowed with drug- like features and to establish a robust pipeline for the fast selection and design of G- quadruplex modulators of emerging bacterial outbreaks based on their sequence”.
Stefano Ciaco collaborates with the research group coordinated by the professor Mattia Mori at the department of Biotechnology, Chemistry and Pharmacy.
This project has received funding from the European Union’s Horizon 2020 programme under GA No 101106871.